RPS-BLAST 2.2.9 [May-01-2004]
Query= local sequence: yp086
(242 letters)
Database: cdd.v2.01
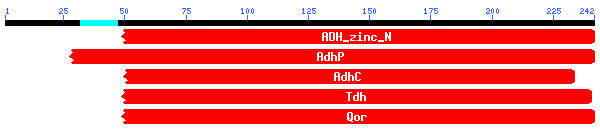
 ..
This CD alignment includes 3D structure. To display structure, download
Cn3D! ..
This CD alignment includes 3D structure. To display structure, download
Cn3D!
| |
PSSMs producing
significant alignments: |
Score
(bits) |
E
value |
| |
 |
gnl|CDD|25397 |
pfam00107, ADH_zinc_N, Zinc-binding
dehydrogenase |
62.4 |
4e-11 |
| |
gnl|CDD|10789 |
COG1064, AdhP, Zn-dependent alcohol
dehydrogenases [General fu... |
134 |
9e-33 |
| |
gnl|CDD|10787 |
COG1062, AdhC, Zn-dependent alcohol
dehydrogenases, class III ... |
46.3 |
3e-06 |
| |
gnl|CDD|10788 |
COG1063, Tdh, Threonine dehydrogenase and
related Zn-dependent... |
39.6 |
3e-04 |
 |
gnl|CDD|10474 |
COG0604, Qor, NADPH:quinone reductase and
related Zn-dependent... |
37.3 |
0.002 |
 |
gnl|CDD|25397, pfam00107, ADH_zinc_N, Zinc-binding
dehydrogenase. | CD-Length = 341 residues, only 62.8% aligned
Score = 62.4 bits (151), Expect = 4e-11
Query: 48 LPEDADPVATAPLLCAGLIGWQSLKMAGE--GRTIGIYGFGAAAHILVQVCKHRGQSVYA 105
Sbjct: 128 IPDGLPLEEAAALGCAGLTAYGALVRAAKVPGDTVLVHGAGGVGLAAIQLAKAAGAAR-V 186
Query: 106 FVLPGDEAGRKFTLDLGAVWAGFSGEKPPVPLDAA----------IIFAPAGELVPMALD 155
Sbjct: 187 IAVDSSEKKLELAKELGAADFVNNSRKEDFVEAIKELTGGKGVDVVLDCVGGATLDAALA 246
Query: 156 VVRKGGTVVCGGI--DMSDIPSMPYRLLWGERRVVSVANLTRSDG------AEFFSIGKA 207
Sbjct: 247 LLKPGGRLVVVGVPGGGAPIPLDPFPLLLKERSIKGSFLGGRKPDELREALDLLASGKGV 306
Query: 208 AGVRCFTSVYPLEHANEALDDLRAGRVSGAAVIVP 242
Sbjct: 307 LKPLITHTLPPLDEAPEAFELMESGKHTGKVVVIP 341
gnl|CDD|10789, COG1064, AdhP, Zn-dependent alcohol
dehydrogenases [General function prediction only] CD-Length = 339 residues, only 65.8% aligned
Score = 134 bits (339), Expect = 9e-33
Query: 27 GYTIDGGFAAHAAAEAGYAFELPEDADPVATAPLLCAGLIGWQSLKMA--GEGRTIGIYG 84
Sbjct: 115 GYTTDGGYAEYVVVPARYVVKIPEGLDLAEAAPLLCAGITTYRALKKANVKPGKWVAVVG 174
Query: 85 FGAAAHILVQVCKHRGQSVYAFVLPGD--EAGRKFTLDLGAVWAG-FSGEKPPVPLDAAI 141
Sbjct: 175 AGGLGHMAVQYAKAMGAEVIAITRSEEKLELAKKLGADHVINSSDSDALEAVKEIADAII 234
Query: 142 IFAPAGELVPMALDVVRKGGTVVCGGI-DMSDIPSMPY-RLLWGERRVVSVANLTRSDGA 199
Sbjct: 235 DTVG-PATLEPSLKALRRGGTLVLVGLPGGGPIPLLPAFLLILKEISIVGSLVGTRADLE 293
Query: 200 EFFSIGKAAGVRC-FTSVYPLEHANEALDDLRAGRVSGAAVIVP 242
Sbjct: 294 EALDFAAEGKIKPEILETIPLDEINEAYERMEKGKVRGRAVIDM 337
gnl|CDD|10787, COG1062, AdhC, Zn-dependent alcohol
dehydrogenases, class III [Energy production and conversion] CD-Length = 366 residues, only 56.0% aligned
Score = 46.3 bits (110), Expect = 3e-06
Query: 49 PEDADPVATAPLLCAGLIGWQSLKMAG---EGRTIGIYGFGAAAHILVQVCKHRGQSVYA 105
Sbjct: 155 DPDAPLEKACLLGCGVTTGIGAVVNTAKVEPGDTVAVFGLGGVGLAAIQGAKAAGAG-RI 213
Query: 106 FVLPGDEAGRKFTLDLGAVWAGFSGEKPPVPL----------DAAIIFAPAGELVPMALD 155
Sbjct: 214 IAVDINPEKLELAKKFGATHFVNPKEVDDVVEAIVELTDGGADYAFECVGNVEVMRQALE 273
Query: 156 VVRKGGTVVCGGI-DMSDIPSMPYRLLWGERRVVSVA---NLTRSDGAEFFSI---GKAA 208
Sbjct: 274 ATHRGGTSVIIGVAGAGQEISTRPFQLVTGRVWKGSAFGGARPRSDIPRLVDLYMAGKLP 333
Query: 209 GVRCFTSVYPLEHANEALDDLRAGRV 234
Sbjct: 334 LDRLVTHTIPLEDINEAFDLMHEGKS 359
gnl|CDD|10788, COG1063, Tdh, Threonine dehydrogenase and
related Zn-dependent dehydrogenases [Amino acid transport and metabolism /
General function prediction only] CD-Length = 350 residues, only 60.3% aligned
Score = 39.6 bits (92), Expect = 3e-04
Query: 48 LPEDADP--VATAPLLCAGLIGWQSLKMAGEGRTIGIYGFGAAAHILVQVCKHRGQSVYA 105
Sbjct: 138 LPDGIDEEAAALTEPLATAYHGHAERAAVRPGGTVVVVGAGPIGLLAIALAKLLGASVV- 196
Query: 106 FVLPGDEAGRKFTLDLGAVWAGFSGEKPPVPL-----------DAAIIFAPAGELVPMAL 154
Sbjct: 197 IVVDRSPERLELAKEAGGADVVVNPSEDDAGAEILELTGGRGADVVIEAVGSPPALDQAL 256
Query: 155 DVVRKGGTVVCGGIDMSDIPSMPYRLLWGERRVV--SVANLTRSD---GAEFFSIGKAAG 209
Sbjct: 257 EALRPGGTVVVVGVYGGEDIPLPAGLVVSKELTLRGSLRPSGREDFERALDLLASGKIDP 316
Query: 210 VRCFTSVYPLEHANEALDDLRAGRVSGAAVIV 241
Sbjct: 317 EKLITHRLPLDDAAEAYELFADRKEEAIKVVL 348
 |
gnl|CDD|10474, COG0604, Qor, NADPH:quinone reductase
and related Zn-dependent oxidoreductases [Energy production and
conversion / General function prediction only] | CD-Length = 326 residues, only 66.3% aligned
Score = 37.3 bits (86), Expect = 0.002
Query: 48 LPEDADPVATAPLLCAGLIGWQSLKMAGE---GRTIGIYGF-GAAAHILVQVCKHRGQSV 103
Sbjct: 111 LPDGLSFEEAAALPLAGLTAWLALFDRAGLKPGETVLVHGAAGGVGSAAIQLAKALGATV 170
Query: 104 YAFVLPGDEAGRKFTLDLGA--------------VWAGFSGEKPPVPLDAAIIFAPAGEL 149
Sbjct: 171 VAVV--SSSEKLELLKELGADHVINYREEDFVEQVRELTGGKGVDVVLDTV-----GGDT 223
Query: 150 VPMALDVVRKGGTVV-CGGIDMSDIPSMPYRLLWGERRVVSVANLTRSDG---------- 198
Sbjct: 224 FAASLAALAPGGRLVSIGALSGGPPVPLNLLPLLGKRLTLRGVTLGSRDPEALAEALAEL 283
Query: 199 AEFFSIGKAAGVRCFTSVYPLEHANEALDDLRA-GRVSGAAVIVP 242
Sbjct: 284 FDLLASGKLKPVI--DRVYPLAEAPAAAAHLLLERRTTGKVVLKV 326
Citing
CD-Search: Marchler-Bauer A, Anderson JB, DeWeese-Scott C,
Fedorova ND, Geer LY, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki
CJ, Liebert CA, Liu C, Madej T, Marchler GH, Mazumder R, Nikolskaya AN,
Panchenko AR, Rao BS, Shoemaker BA, Simonyan V, Song JS, Thiessen PA,
Vasudevan S, Wang Y, Yamashita RA, Yin JJ, and Bryant SH (2003), "CDD:
a curated Entrez database of conserved domain alignments", Nucleic
Acids Res. 31:383-387. |

 ..
This CD alignment includes 3D structure. To display structure, download
Cn3D!
..
This CD alignment includes 3D structure. To display structure, download
Cn3D!